Abstract
Genetic and epigenetic intra-tumoral heterogeneity cooperate to shape the evolutionary course of cancer progression and relapse. In chronic lymphocytic leukemia (CLL), we have previously uncovered a fundamental property of the CLL epigenome: stochastically disordered DNA methylation (DNAme), which reflects ongoing changes in the DNA methylome ('epimutation'). Bulk bisulfite sequencing revealed that DNAme epimutation is significantly higher in CLL compared to normal B cells. Yet, bulk analysis does not allow to phase epimutations across distant genomic loci within the same single cells. As such, it cannot answer whether epimutations occur homogenously across the cell population, or rather affect a minority of cells with highly disordered methylation.
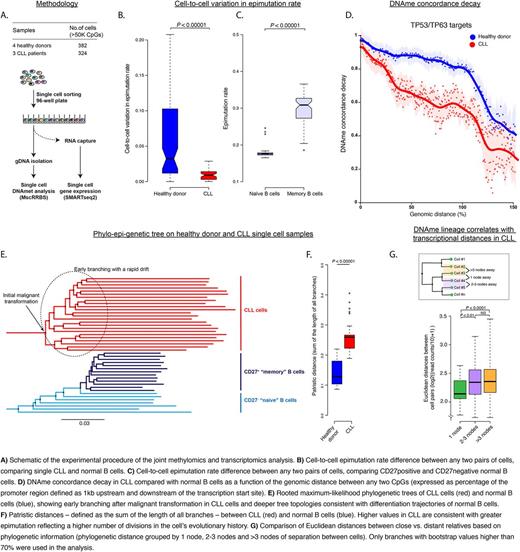
We therefore developed multiplexed single-cell reduced representation bisulfite sequencing (MscRRBS) and applied it to 382 normal B cells from four healthy donors and 324 CLL cells from three CLL patients (Fig A). We achieved bisulfite conversion rates of 99.7%±0.001 (median±SEM) and recovered on average ~500K unique CpG per cell. The overall epimutation rate was higher across CLL cells population compared to single normal B cells, in agreement with our prior bulk RRBS analysis. Uniquely, the single-cell data allowed us to measure cell-to-cell variation in epimutation rate and revealed that CLL samples, irrespective of their IgVH mutational status, exhibit minimal cell-to-cell variability, while normal B cells showed widely variable epimutation rates (Fig B).
This finding is consistent with the hypothesis that epimutation serves as a cellular molecular clock, reflecting the number of generations in the cell's history. Whereas the common clonal CLL origin results in uniform epimutation rate, the variable rate in normal B cells reflects an admixture of cells with different replicative histories. In support of these findings, MscRRBS of sorted naïve and memory B cells revealed aged memory B cells exhibit higher epimutation rate than naïve B cells (Fig C).
Our prior analyses quantified DNAme stochastic epimutation by assessing the concordance of adjacent CpGs recovered within the same sequencing read. As MscRRBS enables the phasing of CpGs even if not contained within the same read, we extended our analysis to calculate the DNAme concordance between any two given CpGs as function of their genomic distance. Consistent with a stochastic epimutation model, we observed faster concordance decay as a function of genomic distance in CLL compared with normal B cells. Indicative of positive selection, this phenomenon was magnified in regions with major roles in gene regulation in CLL (e.g., promoters of tumor suppressor genes such as TP53/TP63 and their targets) (Fig D).
Lastly, the heritable information captured through MscRRBS epimutation data also allowed us to accurately reconstruct phylogenetic relationships between CLL cells, providing a native lineage tracing system. Lineage trees were inferred with maximum likelihood-based phylogeny reconstruction and were found to be 3-fold more robust than simple parsimony trees that ignore the phylogenetic structure. By comparing topological features (maximum tree depth and patristic distance) of normal and CLL phylogenetic trees, we have retraced the evolutionary histories of CLL populations, and found that they exhibit early branching, consistent with rapid drift after the initial malignant transformation. In contrast, normal B cell clades appear to follow a pattern that resembles typical normal B cell differentiation with late branching and deeper tree topology (Fig E, F).
To validate lineage relationships inferred from single-cell DNA methylomes and enable coupling of phylogenetic relationships with transcriptional profiles, we performed parallel MscRRBS and single-cell transcriptome sequencing on 42 individual CLL cells. We found that transcriptional similarity between cells decreases as a function of their phylogenetic distance. Specifically, cells separated by one node (siblings) were ~10% more similar than cells separated by 2 nodes (cousins), providing a direct measurement of the heritability of the transcriptional profile (Fig G). To our knowledge, these measurements offer the first direct comparison of lineage and transcriptional distances between individual cells in primary patient samples, and provide further insight into the epigenetic dimension of CLL evolution.
Wu: Neon Therapeutics: Consultancy. Landau: Pharmacyclics: Membership on an entity's Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal